Theory and Simulation of Biomolecular Interactions and Flexibility
The current understanding of biomolecular interactions that underlie normal and aberrant sub-cellular processes is limited. The role of these interactions in cancer and in a host of viral- and bacterial-caused diseases is even less well understood at the atomistic level. Many biomolecular interactions occur at the level of post-transcriptional gene regulation involving intricate networks of interacting protein and RNA molecules. The research in our group focuses on the application and development of theoretical and computational methods with the intent of gaining an in-depth understanding of biological interactions and functions. In these endeavors, we uses simulation based approaches, and related statistical mechanics, classical and quantum mechanical methods.
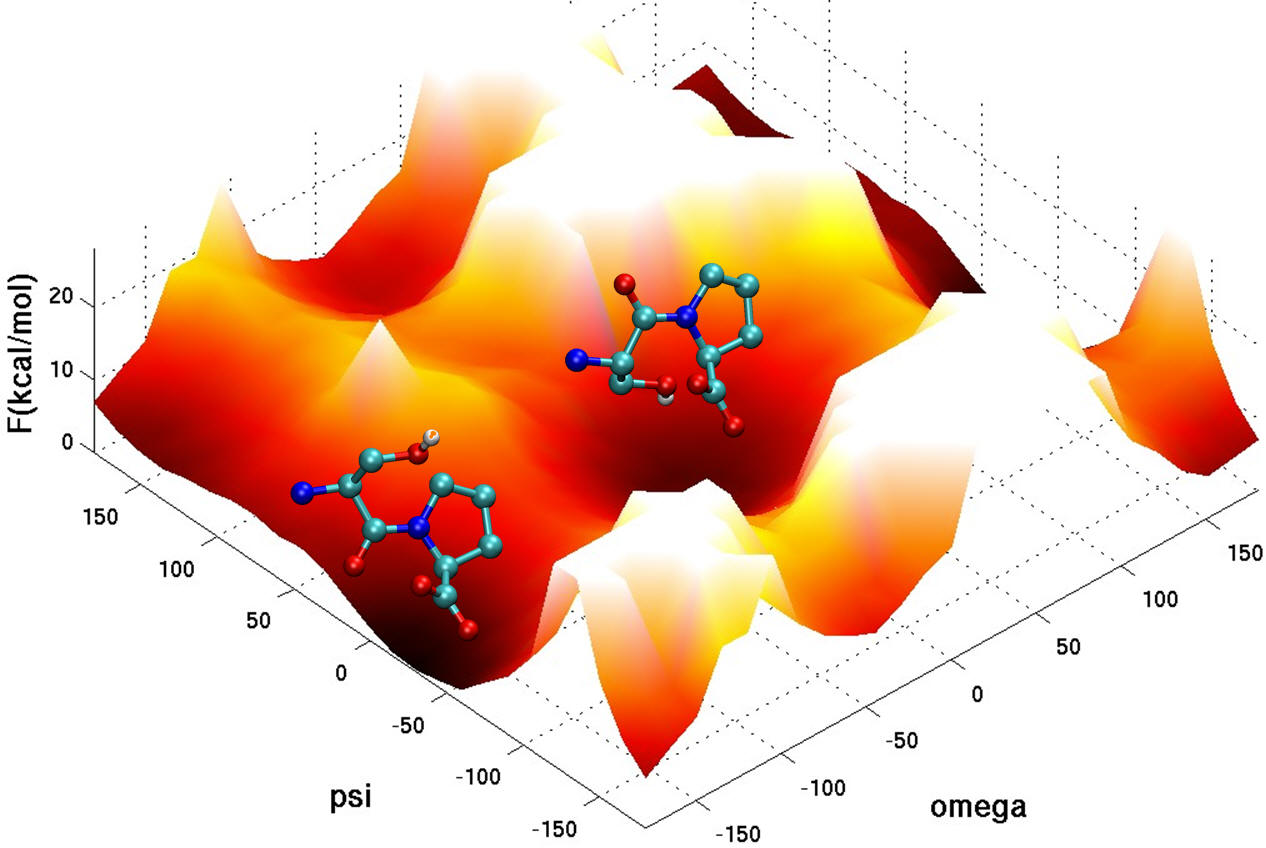
We also use advanced simulation methods to study the dynamic fluctuations of biomolecules. Mechanisms of biomolecular interaction are not as simple as a rigid lock-and-key model, but are rather modulated and optimized by some amount of flexibility. In order to perform their catalytic activities, enzymes generally have to undergo structural transitions upon interaction with other proteins or ligands and upon release of products. The opening and closing of the flaps that act as gates to the active site of HIV-1 protease is a recognizable example. Also, Conformational transitions in one part of a protein can modulate the affinity of its binding partners due to changes in the structural features within the binding site.There must therefore be some form of structural "communication" between the distant sites, and this necessitates an explanation that in some way invokes dynamics.
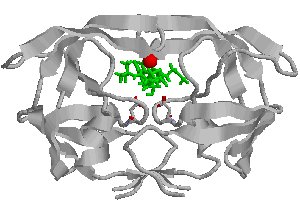
Furthermore, the general understanding of hydration effects at binding sites and interfaces in biomolecular interactions is also of interest in our group. Water molecules make up an integral part of protein structures, while assisting in catalysis, providing stability and controlling the plasticity of binding sites. They are characteristically present in the interfaces found in protein-ligand, protein-protein, protein-carbohydrate, protein-nucleic acid, and nucleic acid-ligand complexes. We use MD-based simulation techniques and free energy calculations to gain insight into the nature of these water molecules.
The research in our group benefits from locally available Supercomputer and compute cluters operated by Research Computing at Georgia State University and network of high performance computers in our group.